Input
- Reference accesion Number :
- - In order to run the BLAST with query contigs and reference, Reference sequences are required. For more information, please refer to NCBI site
- Input File Format :
- - Input file is the assembled contigs and format should be in FASTA format. The number of contigs are not limimted, but, it will takes several hours. We recommend to copy the reslut page URL for your reference.
- Maximum number of matched BLAST hit group :
- - The number sets the maximum groups which is based on the BLAST result.
- Minimum matched sub gene's count per each contig :
- - i Please refer to below Figure 1-B.
Option value
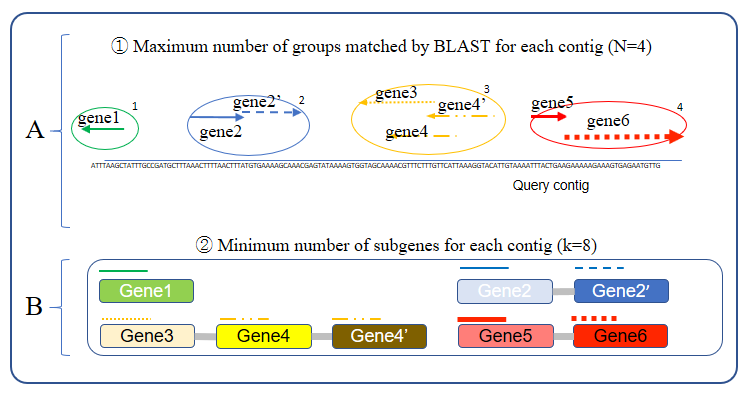
Figure 1. Maximum number of hit group and mininum number of subgene's count
Output
- Sorted by the number of subgene :
- BLAST result file which is sorted by BLAST e-value. In the sorted query sequences file, sequences are sorted by the number of matched sub-genes and the sequences that do not meet the minimum value of k are filtered out
- PDF file:
- The results are shown as a graph depicting matches with the 149 reference genomes on the X-axis and query sequences uploaded by the user on the Y-axis. Each 150 line on the plot indicates that a query contig is matched with the reference sequences.